|
Focused Ion Beam (FIB) Scanning Electron Microscopy (SEM)
by Hall, Hartwieg and Nguyen
doi:10.3908/wormatlas.9.16
Description
One of the shortcomings of serial thin sections (40–80 nm thick) is the relatively poor resolution of structures along the Z-axis compared to 1–2 nm resolution obtained in both X-and Y-axes. Electron tomography has helped to emphasize this difference, since it has allowed us to see smaller cytoskeletal elements (say 535 nm) that might be hiding behind one another in a thin section, but which become individually separable after tilting the section. However, electron tomography has rather limited throughput, and is currently only practical within relatively small volumes of tissue (one cell at a time), and perhaps not suitable for resolving all cells and organelles at once in the whole animal. One alternative solution to this dilemma is to shave through a plastic-embedded sample at much thinner steps (510 nm or less), discarding the sectioned tissue, and collecting serial images of the block face as it is progressively eroded. High-energy focused ion beams (FIB) can accomplish tissue erosion in a controlled fashion within the vacuum chamber of a scanning electron microscope, that is, a FIB/SEM (Knott et al., 2008; Bushby et al., 2012). Between each erosion step, the SEM is used in back-scatter mode to view the new block-face, collecting new electron microscopic image(s) of the newly revealed surface features at a depth of 20-50 nm. By serially eroding the block and imaging the block-face, a serial movie can be produced through the sample.
Similar to the block-face imaging approach of the Denkotome (see Serial block-face SEM), FIB/SEM offers a theoretical advantage over conventional serial section imaging due to much better image registration. Thin sections are subject to warping, wrinkling and chatter that interfere with the production of a glitch-free movie through the tissue. As the resolution of block-face imaging improves, this technology holds great promise for the accurate reconstruction of small volumes of tissue at a resolution of 5 nm or better (EMFIBSEMFIG 1 and 2).
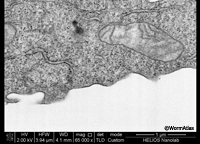 EMFIBSEMFIG 1 EMFIBSEMFIG 1 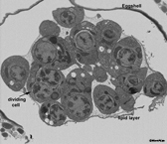 EMFIBSEMFIG 2 EMFIBSEMFIG 2
The FIB/SEM method has been limited by technical difficulties in masking the sample block from degradation by the ion beams, sheltering the block face from contamination during erosion, and obtaining high-resolution images of the new block face, using backscattered electrons to peer into the surface of the block. But technical progress is being made to improve the tissue staining, thus enhancing visibility of tissue elements (see OTO Fixation Method). In the same manner as for the 3View SEM (see Serial block-face SEM), fine control of the accelerating voltages should help to limit the zone of electron imaging to a relatively shallow depth to match the narrow erosion steps and achieve finer resolution along that axis. EMFIBSEMFIG 3 shows an example from current work (in 2010) using FIB/SEM to explore nematode anatomy in three dimensions.
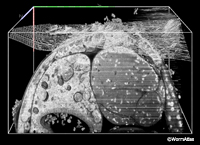 EMFIBSEMFIG 3 EMFIBSEMFIG 3
In the near future, we expect that FIB/SEM instruments will become much more common in biological studies of the ultrastructure, and that improvements in resolution will permit this technology to replace serial thin section techniques for certain ambitious projects, such as imaging whole C. elegans embryos at different developmental stages. Current vendors producing FIB/SEM devices include Zeiss, FEI (EMFIBSEMFIG 4) and TESCAN. However, slow throughput with these devices makes it generally impractical to reconstruct whole animals at very high resolution (see Drawbacks section below).
 EMFIBSEMFIG 4 EMFIBSEMFIG 4
EMFIBSEMMOV 1 shows a video produced from images acquired on a FEI Helios FIB/SEM microscope of a wild type L1 head milled in 10nm through the nerve ring.
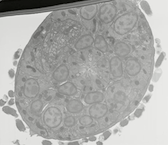 EMFIBSEMMOV 1 EMFIBSEMMOV 1
Troubleshooting
As the capability of the FIB/SEM method is improving, its sensitivity to the building environment is becoming more acute. When trying to mill away only 510 nm per step, the timing cycle between each milling step and the imaging step may become critical (so that the block remains stable in the long term with no local heating or warping of the block itself), and the device may require extreme isolation from building vibrations, temperature variations, ventilation drafts, etc.
Drawbacks
Given the large number of images required when one shaves the tissue in such thin sections, the total time to acquire all images can also become a drawback, since one then requires even greater amounts of time per sample. This method destroys the tissue sample as it progresses, so there is no chance to re-shoot any portion of the image series later. All aspects of the imaging must be correct on the first attempt. It can be challenging to know when to start image collection, since the specimen lies hidden within an opaque mass prior to sectioning. Therefore the operator must begin imaging before the critical portion of the animal has been reached.
Acknowledgements
We thank William Rice and Ruben Diaz-Avalos at the New York Structural Biology Center (NYSBC) for their help in exploring the capabilities of FIB/SEM on C. elegans samples (EMFIBSEMFIG 2 and MOV 1). We also thank Jemima Burden (University College London) for sharing the images shown in EMFIBSEMFIG 1 and 3.
Figures
 Click pictures for new window with figure and legend, click again for high resolution image Click pictures for new window with figure and legend, click again for high resolution image
EMFIBSEMFIG 1: FIB/SEM imaging of cultured cell. High-power view of block-face image of a cultured cell after FIB erosion, FEI Helios FIB/ SEM. Work of Ian White and Jemima Burden (University College, London) and David Wall (FEI).
EMFIBSEMFIG 2: FIB/SEM imaging of C. elegans embryo. Wild type C. elegans embryo at the cell proliferation stage is viewed by focused ion beam milling and SEM. Tissues are not yet formed, but early blastomeres are viewed at different cell cycle stages. Most of these will later give rise to multiple cell types. Gastrulation has begun, and some cells are now internalized. A few cells extend pseudopodia, either to facilitate local cell movements, or perhaps as early steps in morphogenesis. Substantial extracellular space separates the embryonic cells from the eggshell, while a distinct lipid layer encloses all of the cells more tightly. Fluid beneath the lipid layer is destined to become the early basis for the blastocoel.
Specimen was processed by Ken Nguyen using high pressure freezing, freeze substitution, and an OTO protocol, before infiltration into plastic resin. Blockface imaging by SEM was conducted by Dr. William Rice using the FEI Helios FIB/SEM at New York Structural Biology Center.
EMFIBSEMFIG 3: FIB/SEM imaging. Three-dimensional model in C. elegans where the data has been digitally resectioned to view from three different angles within the cube of tissue (bounding box is 60 x 30 x 30 mm in volume). Shown are the intestine and its lumen, and the proximal gonad arm in the adult hermaphrodite midbody. Some noise is evident from the serial SEM images, which were acquired over 12.5 h, FEI Quanta 3D ESEM. Work of Rachel Ward and Jemima Burden, University College, London, and Ken Png and Andy Bushby, The Nanovision Centre, Queen Mary University of London.
EMFIBSEMFIG 4: FEI Helios Nanolab 650. A. FEI Helios Nanolab 650 focused ion beam milling SEM used for acquiring FIB/SEM data. B. Closeup of the specimen cryo-transfer station for the Helios 650 FIB/SEM, which permits frozen samples to be transferred into the chamber for focused ion beam milling.
EMFIBSEMMOV 1: FIB/SEM movie of L1 nerve ring. Wild type L1 head was milled in 10 nm steps through a portion of the nerve ring. The animal is rotated so that ventral axis is to the upper left corner. The first image begins in the retrovesicular ganglion, and the movie proceeds anteriorly through the entire nerve ring. Tissue was fixed using an OTO protocol by Ken Nguyen, and images were acquired on the FEI Helios FIB/SEM microscope at the New York Structural Biology Center by Dr. William Rice. Use buttons under image to play and pause embedded movie. Click on image to open movie in a new window.
References
Bushby, A.J., Mariggi, G., Armer, H.E. and Collinson, L.M. 2012. Correlative light and volume electron microscopy: using focused ion beam scanning electron microscopy to image transient events in model organisms. Methods Cell Biol. 111: 357-82. Abstract
Hall, D.H., Hartweig, E. and Nguyen, K.C.Q. 2012. Modern electron microscopy methods for C. elegans. Methods Cell Biol. 107: 93-149. Abstract
Knott, G., Marchman, H., Wall, D. and Lich, B. 2008. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J. Neurosci. 28: 2959–2964. Article
|
