

|
The Evidence for Classical Neurotransmitters in C. elegans Neurons
This table covers the classical small molecule neurotransmitters for which there is substantial published evidence that they are present, and are likely to function in specific C. elegans neurons. This evidence falls under 'criterion I' as outlined in Criteria for assigning a neurotransmitter function in C. elegans. Other molecules that may serve as neurotransmitters in C. elegans, but for which there is little or no published evidence, are generally not included here. This table also does not typically include substances released by non-neuronal cells that may act in a hormonal or transmitter-like function. [See Hobert (2013) for a presentation of 'the case for and against other neurotransmitter systems' and other transmitter-related aspects of the 'neuronal genome.'] See also Pereira et al. (2015), Gendrel et al. (2016) and Serrano-Saiz et al., 2017 for extensive treatments of neurotransmitters in C. elegans, and the Hobert Lab Neurotransmitter Map Resource.
by Curtis M. Loer and James B. RandIn the literature cited, there are occasional disagreements about which specific cells show expression in reporter fusion transgenics. We have included in this table published cell "identifications" even for cases in which there was a likely mis-identification. (We have attempted to identify such instances in the footnotes.) In some cases, the situation is less clear, and could reflect real expression differences arising from reporter constructs with different regions of sequence, integration sites, etc. Each Summary List of neurons represents our best judgment based on the evidence available. In general, when conflicts arise, we defer to studies that are 1) more recent, 2) comprehensive, and/or 3) use constructs more likely to represent the true expression pattern (e.g., fosmid or single-copy insertions into native loci).
Acetylcholine (ACh)
Biogenic Amines
Dopamine
Tyramine (TA) / Octopamine (OA)
Serotonin (5HT)
Gamma-aminobutyric acid (GABA)
Glutamate (Glu)Neurons currently lacking classical neurotransmitter assignments
Footnotes
Abbreviations
Acknowledgments and General References
How to cite this documentAll Neurons with Neurotransmitter IDs - Excel spreadsheet [now updated from the previous WormAtlas 2016 version]
[Note: The neuron list comes originally from Individual Neurons on WormAtlas; the tables are mostly derived with permission from the tables in Pereira et al. (2015) and Serrano-Saiz et al., 2017 supplemental materials.]
| ACETYLCHOLINE (ACh) | ||||||
| Summary List of ACh neurons | ||||||
| DESCRIPTION | GENE NAME | DETECTION METHOD | LOCALIZATION | REFERENCES | ||
| ACh | NA | Radioenzymatic assay | Whole animal | Hosono et al., 1987; Hosono & Kamiya, 1991; Nguyen et al., 1995 | ||
Synthesis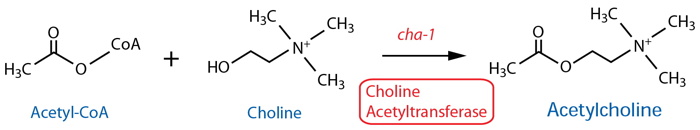 |
||||||
| Choline Acetyltransferase (ChAT) | cha-11 |
Enzymatic assay | Whole animal | Rand & Russell, 1984 | ||
| ChAT | cha-11 |
Antibody | Same as unc-17 (see below)2 | Duerr et al., 2008 | ||
| Transport | ||||||
| Vesicular Acetylcholine Transporter (VAChT) | unc-171 |
Antibody | AIA, AIY | Altun-Gultekin et al., 2001 | ||
| VAChT | unc-171 |
Antibody | ALN, AS1-11, DA1-9, DB1-7, HSN (faint, h), PLN, SDQ, URA, URB, VA1-12, VB1-11, VC1-6(h), many others | Duerr et al., 2008 | ||
| VAChT | unc-171 |
Reporter transgenics | IL2, URA, URB | Zhang et al., 2014 | ||
| VAChT | unc-171 |
Reporter transgenics | PCB(m), PCC(m), PVX(m), PVY(m), SPC(m), SPV(m) | Garcia et al., 2001; LeBouef et al., 2014 | ||
| VAChT | unc-171 |
Reporter transgenics | SMB | Kim et al., 2015 | ||
| VAChT | unc-171 |
Reporter transgenics (fosmid3), Antibody | ADF, AIA, [AIM]4, AIN, AIY, ALN, AS1-11(1-11), ASJ, AVA, AVB, AVD, AVE, AVG5, AWB, CA1-9(m), CEM(m), DA1-9, DB1-7, DVA6, DVE(m)7, DVF(m)7, HOB(m), HSN(h), I15, I35, IL2, M15, M25, M4, M5, MC5, PCB(m), PCC(m), PDA, PDB, PDC(m), PGA(m), PLN, PVC, PVN5, PVP, PVV(m), PVX(m), PVY(m), PVZ(m), R1A(m), R2A(m), R3A(m), R4A(m), R6A(m), RIB8, RIF, RIH, RIR, RIV, RMD, RMF, RMH, SAA5, SAB, SDQ, SIA, SIB, SMB, SMD, SPC(m), SPV(m), URA, URB, URX, VA1-12, VB1-11, VC1-6(h)9 | Pereira et al., 2015 | ||
| VAChT | unc-171 | Reporter transgenics (fosmid3) | DX1/2(m)7 | Serrano-Saiz et al., 2017 | ||
| Choline Transporter (ChT) |
cho-1 |
Reporter transgenics | Most cholinergic (i.e., unc-17-expressing) neurons | Okuda et al., 2000; Matthies et al., 2006; Mullen et al., 2007 | ||
| ChT | cho-1 | Reporter transgenics | IL2, URA | Zhang et al., 2014 | ||
| ChT | cho-1 | Reporter transgenics (fosmid3) | Expressed in all unc-17-expressing neurons EXCEPT AVG, CA7-9(m), DX1/2(m)7, HSN(h), I1, I3, M1, M2, MC, PVN, SAA, and VC4-5(h); also expressed in R8A(m), R8B(m) | Pereira et al., 2015; Serrano-Saiz et al., 2017 | ||
| ChT | cho-1 | Uptake assay10 | NA - heterologous expression (Xenopus oocyte) | Okuda et al., 2000 | ||
| Postsynaptic Choline/Acetylcholine Transporter | snf-6 |
Reporter transgenics | Neuromuscular junctions of body wall muscle, vulval and enteric muscles; a few unidentified neurons | Kim et al., 2004 | ||
Catabolism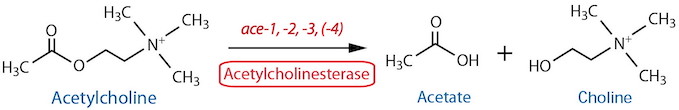 |
||||||
| Acetylcholinesterase (AChE) Total | NA | Histochemistry11 | Strong, reliable staining in nerve ring, ventral ganglion, pharyngeal-intestinal valve and anal depressor region; more variable staining in VNC, DNC and PAG | Culotti et al., 1981 | ||
| AChE class A | ace-1 |
Enzymatic assay | Whole animal | Johnson et al., 1981 | ||
| AChE class B | ace-2 |
Enzymatic assay | Whole animal | Johnson et al., 1981; Culotti et al., 1981 | ||
| AChE class C | ace-3 |
Enzymatic assay | Whole animal | Kolson et al., 1985a; Kolson et al., 1985b; Johnson et al., 1988 | ||
| AChE | ace-1 |
Reporter transgenics | CEP, OLL, pm5, body wall muscles, vulval muscles, anal sphincter muscle | Culetto, 1999 | ||
| AChE | ace-1 |
Reporter transgenics (fosmid3) | CEP, OLL | Pereira et al., 2015 | ||
| AChE | ace-2 |
Reporter transgenics | IL cells, AWB(?), AWC, additional head neurons, pm5, PVC, PVQ, PDA, hyp 8-11 | Combes et al., 2003 | ||
| AChE | ace-2 |
Reporter transgenics (fosmid3) | AS1-11, AVA, AVB, AVD, AVE, DA1-9, DB1-7, DVA, M4, PDA, RIH, VA1-1, VB1-11, others | Pereira et al., 2015 | ||
| AChE | ace-3/ace-412 |
Reporter transgenics | pm3, pm4, pm5, pm7, CAN, some body muscles | Combes et al., 2003 | ||
| AChE | ace-3/ace-412 | Reporter transgenics | AIA, DVA, IL2, PDA, RIH, RIV, RMD, SIA, SMD, URA, URB, URX, others | Pereira et al., 2015 | ||
| Related Mutant Phenotypes / Other Supportive Evidence | ||||||
| ACh | cha-1 |
ACh levels reduced or absent | Hosono et al., 1987 | |||
| ACh | unc-17 |
ACh levels elevated | Hosono et al., 1987 | |||
| ChAT | cha-1 |
ChAT enzymatic activity is reduced or absent; hypomorphic cha-1 mutants are Unc and Ric (resistant to inhibitors of cholinesterase); null mutants arrest shortly after hatching (lethal). | Rand & Russell 1984; Hosono et al., 1987; Rand, 1989 | |||
| VAChT | unc-17 |
Hypomorphic unc-17 mutants are Unc and Ric; null mutants arrest shortly after hatching (lethal). | Brenner, 1974; Rand & Russell 1984; Alfonso et al., 1993 | |||
| VAChT | unc-17 |
Mutant analysis indicates cholinergic function in pharyngeal neuron MC. | Raizen et al., 1995 | |||
| VAChT | unc-17 |
Mutant analysis indicates cholinergic function in neuron IL2. | Lee et al., 2012 | |||
| AChE | ace-1 | Class A - AChE activity is absent. ace-1 mutants have no behavioral phenotype. | Johnson et al., 1981 | |||
| AChE | ace-2 | Class B - AChE activity is absent. Histochemical staining4 is reduced in ace-2 mutants, and completely eliminated in ace-2; ace-1 double mutants. ace-2 mutants are hypersensitive to Aldicarb, but have no behavioral phenotype, but ace-2; ace-1 double mutants are Unc. | Culotti et al., 1981 | |||
| AChE | ace-3 | Class C- AChE activity is absent. ace-3 mutants have no behavioral phenotype; ace-3; ace-1 and ace-2 ; ace-3 double mutants have ~wild type behavior; ace-2 ; ace-3; ace-1 triple mutants arrest as L1s (lethal). | Johnson et al., 1988 | |||
SUMMARY - Cholinergic Neurons (adult): Hermaphrodite (n = 160), Male (n = 193) By Class (with numbers and notes):
By Body Region: In all Summary lists, By Body Region - Somas found in the retrovesicular ganglion (RVG) are listed as head neurons; those in the preanal ganglion (PAG) are listed as tail neurons. | ||||||
| BIOGENIC AMINES | ||||||
| DOPAMINE (3-hydroxytyramine, dihydroxyphenylethylamine) | ||||||
| Summary List of Dopaminergic Neurons | ||||||
| DESCRIPTION | GENE NAME | DETECTION METHOD | LOCALIZATION | REFERENCES | ||
| Dopamine | NA | HPLC + ED | Whole animal | Sanyal et al., 2004 | ||
| Dopamine | NA | Formaldehyde induced fluorescence (FIF) | CEP, ADE, PDE, R5A(m), R7A(m), R9A(m) | Sulston et al., 1975 | ||
Synthesis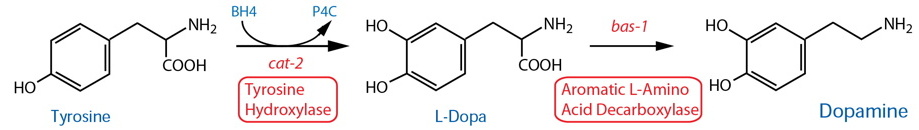 |
||||||
| Tyrosine Hydroxylase (TH) | cat-2 |
Reporter transgenics | CEP, ADE, PDE, R5A(m), R7A(m), R9A(m), |
Lints & Emmons, 1999; Flames & Hobert, 2009 ; LeBouef et al., 2014 | ||
| Aromatic L-Amino Acid Decarboxylase (AADC)17 | bas-1 |
Reporter transgenics | CEP, ADE, PDE, R5A(m), R7A(m), R9A(m), |
Hare & Loer, 2004; Flames & Hobert, 2009 ; LeBouef et al., 2014 | ||
BH4 Cofactor Synthesis and Regeneration
|
||||||
| GTP |
cat-4 |
Reporter transgenics | CEP, ADE, PDE, R5A(m), R7A(m), R9A(m); see also 5HT neurons | Sze et al., 2002; Flames & Hobert, 2009 | ||
| GTPCH1 | cat-4 | Enzymatic assay | Whole animal | Loer et al., 2015 | ||
| Pyruvoyl Tetrahydropterin Synthase (PTPS) | ptps-116 |
Reporter transgenics | Various unidentified neurons and non-neuronal cells; see also 5HT neurons | Zhang et al., 2014; Loer et al., 2015 | ||
| PTPS | ptps-1 | Enzymatic assay | Whole animal | Loer et al., 2015 | ||
| Pterin Carbinolamine Dehydratase (PCBD) | pcbd-116 |
Reporter transgenics | Various unidentified neurons and non-neuronal cells; see also 5HT neurons | Zhang et al., 2014; Loer et al., 2015 | ||
| Quinoid Dihydropterin Reductase (QDPR)17 | qdpr-116 |
Reporter transgenics | CEP, other unidentified cells; see also 5HT neurons | Zhang et al., 2014; Loer et al., 2015 | ||
| Transport | ||||||
| Vesicular Monoamine Transporter (VMAT) | cat-1 |
Antibody19 | CEP, ADE, PDE; see also 5HT, OA, and TA neurons | Duerr et al., 1999 | ||
| VMAT | cat-1 |
Reporter transgenics | CEP, ADE, PDE, R5A(m), R9A(m); see also 5HT, OA, and TA neurons | Flames & Hobert, 2009 | ||
| VMAT | cat-1 |
Reporter transgenics (fosmid3) | CEP, ADE, PDE, R5A(m), R7A(m), R9A(m); see also 5HT, OA, and TA neurons | Serrano-Saiz et al., 2017 | ||
| VMAT | cat-1 |
Uptake assay21 | NA - heterologous expression (CV-1 cells) | Duerr et al., 1999 | ||
| (Plasma Membrane) Dopamine Transporter (DAT) | dat-1 |
Reporter transgenics | CEP, ADE, PDE, R5A(m), R7A(m), R9A(m) | Nass et al., 2001; Nass et al., 2002; Flames & Hobert, 2009 | ||
| DAT | dat-1 | Antibody | CEP, ADE, PDE(rare) | McDonald et al, 2007 | ||
| DAT | dat-1 |
Uptake assay22 | NA - heterologous expression (HeLa cells) | Jayanthi et al., 1998 | ||
| DAT | dat-1 |
Uptake assay, patch clamping recording23 |
Heterologous expression (tsA-201 cells) and cultured embryonic C. elegans dopaminergic neurons (Pdat-1::GFP cells) | Carvelli et al., 2004 | ||
Catabolism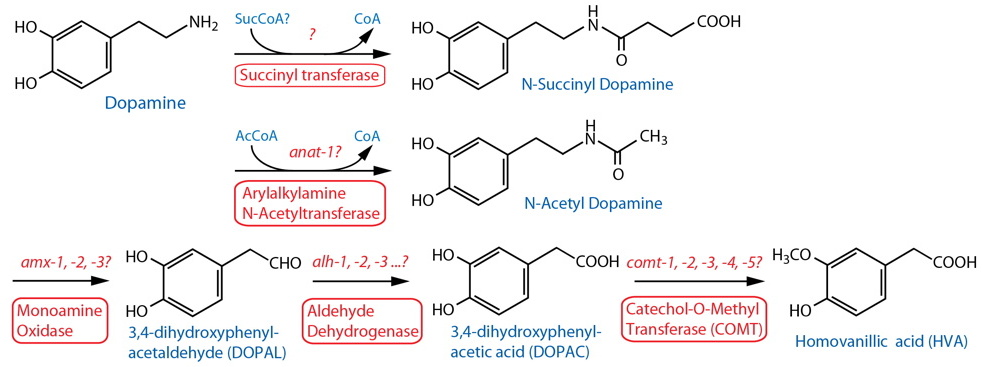 [Note: Monoamine catabolism pathways are only partially characterized in C. elegans. N-acetylation and N-succinylation are likely significant destinations for monoamines including dopamine (Artyukhin et al., 2013). Oxidation via AMX-2/MAO is likely, similar to the well-characterized pathways in other animals (best known from vertebrates, especially mammals). There are no clear orthologs of catechol-o-methyl transferase (COMT); however, worm comt genes encode proteins with a methyltransferase associated domain also found in mammalian COMT24. Several different monoamine inactivating modifications are found among invertebrate phyla, including N-acetylation, γ-glutamylation, β-alanylation, sulfation, etc. (see review by Sloley, 2004), although it is not always clear whether these are associated with neurons, or that the modifications are catabolic in nature.] |
||||||
| Arylalkylamine N-Acetyltransferase (AA-NAT) |
anat-1? | Enzymatic assay | Whole animal | Migliori et al., 2012 | ||
| Monoamine Oxidase (MAO) | amx-125 |
Reporter transgenics | ~30 head and tail neurons including ASJ, IL2, other amphid neurons; PHA, PHB, 3 other tail neurons; not expressed in dopaminergic cells. Expressed in nearly all cells in the embryo. | Filkin et al., 2007; Kaushal 2008 | ||
| MAO | amx-225 |
Reporter transgenics | 4 pairs amphid neurons: ASJ, and most likely ASH, ASE, AWB, PHA, PHB | Hostettler et al., 2017 | ||
| MAO | amx-225 |
Reporter transgenics | Intestine, neurons | Filkin et al., 2007 | ||
Aldehyde Dehydrogenase (ALDH) |
alh-126 | Reporter transgenics | Nervous system, including head neurons, neurons along body, PVT, intestine, head mesodermal cell, rectal gland cells, etc. | McKay et al., 200327 | ||
ALDH |
alh-626 | Reporter transgenics | Body wall muscle, hypodermis, unidentified cells in head, unidentified cells in tail (adult) | McKay et al., 200327 | ||
Succinic Semialdehyde Dehydrogenase (SSADH) |
alh-726 | Reporter transgenics | Intestine, rectal gland cells, nervous system, head neurons (adult) | McKay et al., 200327 | ||
ALDH |
alh-1026 | Reporter transgenics | Intestine, nervous system, tail neurons (adult) | McKay et al., 200327 | ||
| Related Mutant Phenotypes / Other Supportive Evidence | ||||||
| Dopamine | cat-1, cat-2, cat-4 | Dopamine levels (by HPLC + ED) are reduced to about 40% of wildtype in each of these three mutants. | Sanyal et al., 2004 | |||
| TH | cat-2 | Mutant lacks dopamine by FIF; see also 5HT-related phenotypes. | Sulston et al., 1975 | |||
| AADC | bas-1 | Mutant lacks dopamine by FIF; Dopaminergic cells do not become serotonin-immunoreactive with 5HTP treatment; see also 5HT-related phenotypes. | Sawin et al., 2000; Loer & Kenyon, 1993 | |||
| AADC | bas-1 | Age-related decline in bas-1 mRNA levels and reporter expression correlate with reduced FIF in CEPs. | Yin et al., 2014 | |||
| GTPCH1 | cat-4 | Mutant lacks dopamine by FIF; see also 5HT-related phenotypes. | Sulston et al., 1975; Desai et al., 1988; Loer et al., 2015 | |||
| GTPCH1 | cat-4 | Mutant lacks GPCH1 enzymatic activity. | Loer et al., 2015 | |||
| PTPS | ptps-1 | Mutant lacks dopamine by FIF; see also 5HT-related phenotypes. | Loer et al., 2015 | |||
| PTPS | ptps-1 | Mutant lacks PTPS enzymatic activity. | Loer et al., 2015 | |||
| QDPR | qdpr-1 | Mutant has reduced dopamine by FIF, especially combined with cat-4 reduction-of-function mutation; see also 5HT-related phenotypes. | Loer et al., 2015 | |||
| PCBD | pcbd-1 | Mutant has reduced dopamine by FIF, especially combined with cat-4 reduction-of-function mutation; see also 5HT-related phenotypes. | Loer et al., 2015 | |||
| VMAT | cat-1 | Dopamine by FIF in mutant is reduced in processes and increased in somas; mutant is phenocopied by reserpine (VMAT blocker). | Sulston et al., 1975 | |||
| VMAT | cat-1 | Expression of human VMAT1 or VMAT2 protein in mutant partially rescues behavioral phenotypes, and restores 5HT & dopamine-induced fluorescence (GAIF). | Duerr et al., 1999 | |||
| VMAT | cat-1 | Null mutants are deficient in dopamine-mediated behaviors. | Duerr et al., 1999 | |||
| DAT | dat-1 | Null mutants lack dopamine uptake in cultured embryonic dopaminergic cells. | Carvelli et al., 2004 | |||
| DAT | dat-1 | Mutants are protected from 6-OHDopamine-induced dopaminergic neuron degeneration, consistent with role of DAT-1 in dopamine uptake. | Nass et al., 2001; Nass et al., 2005 | |||
| DAT | dat-1 | Mutants display 'swimming induced paralysis' (SWIP) phenotype consistent with excess extrasynaptic dopamine; SWIP is reduced by pretreatment with reserpine, and abolished in cat-2; dat-1 double mutant. | McDonald et al., 2007 | |||
SUMMARY - Dopaminergic Neurons (adult): Hermaphrodite (n = 8), Male (n = 14+2)13 By Class (with numbers and notes): By Body Region: |
||||||
TYRAMINE (TA) and OCTOPAMINE (OA) |
||||||
| Summary List of TA & OA Neurons | ||||||
| DESCRIPTION | GENE NAME | DETECTION METHOD | LOCALIZATION | REFERENCES | ||
| TA | NA | TLC | Whole animal | Alkema et al., 2005 | ||
| OA | NA | Radioenzymatic assay 28 | Whole animal | Horvitz et al., 1982 | ||
| OA | NA | HPLC + ED | Whole animal | Alkema et al., 2005 | ||
Synthesis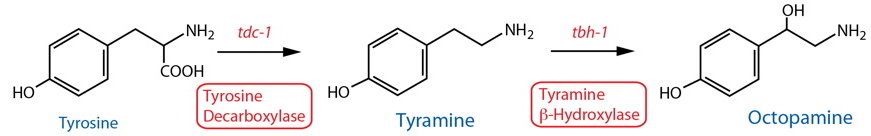 |
||||||
| Tyrosine Decarboxylase (TDC) | tdc-1 |
Enzymatic assay | Whole animal | Alkema et al., 2005 | ||
| TDC | tdc-1 |
Reporter transgenics, Antibody | RIC, RIM, uv129, gonadal sheath cells | Alkema et al., 2005 | ||
| TDC | tdc-1 |
Reporter transgenics | RIC, RIM, HOA(m), R8A(m)30, R8B(m)30 | Serrano-Saiz et al., 2017 | ||
| Tyramine Beta-Hydroxylase (TBH) | tbh-1 |
Reporter transgenics, Antibody | RIC, gonadal sheath cells | Alkema et al., 2005; Suo et al., 2006 | ||
| Transport - TA and OA are both likely substrates for transport by VMAT | ||||||
| Vesicular Monoamine Transporter (VMAT)18 | cat-1 | Reporter transgenics | RIC; see also dopaminergic, 5HT neurons | Duerr et al., 1999 | ||
| VMAT | cat-1 |
Reporter transgenics (fosmid3) | RIC, RIM, HOA(m); see also dopaminergic, 5HT neurons | Serrano-Saiz et al., 2017 | ||
| VMAT | cat-1 |
Antibody19 | RIC; see also dopaminergic, 5HT neurons | Duerr et al., 1999 | ||
| VMAT | cat-1 |
Uptake assay21 | TA and OA are competitive inhibitors of dopamine and 5HT uptake by CAT-1 in heterologous expression (CV-1 cells). | Duerr et al., 1999 | ||
Catabolism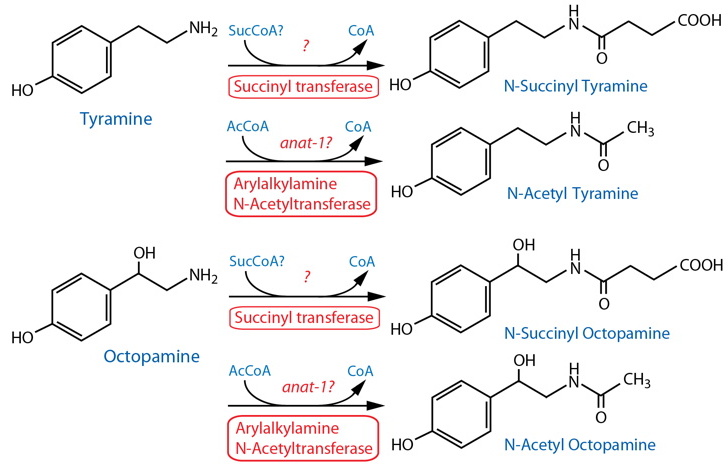 [Note: Monoamine catabolism pathways are only partially characterized in C. elegans. Succinylation appears to be a significant pathway for TA and OA (Artyukhin et al., 2013). TA and OA may also be catabolized via MAO, likely AMX-2. See Dopamine catabolism for further notes on possible monoamine metabolism, and lists of genes that may be involved.] |
||||||
| Related Mutant Phenotypes / Other Supportive Evidence | ||||||
| TA | tdc-1 | Mutant lacks TA (by TLC). | Alkema et al., 2005 | |||
| TA, OA | tdc-1 | Mutant lacks both tyramine- and octopamine succinyl ascarosides. | Artyukhin et al., 2013 | |||
| TA, OA | tbh-1 | Mutants lacks octopamine succinyl ascarosides; excess succinylated derivative of tyramine is produced. | Artyukhin et al., 2013 | |||
| OA | tdc-1, tbh-1 | Mutants lack OA (by HPLC+ ED). | Alkema et al., 2005 | |||
| TDC | tdc-1 | TDC enzymatic activity is absent or strongly reduced. | Alkema et al., 2005 | |||
SUMMARY - Tyraminergic Neurons (adult): Hermaphrodite (n = 2), Male (n = 3-730) By Class (with numbers and notes): By Body Region: |
||||||
| SUMMARY - Octopaminergic Neurons (adult): Hermaphrodite (n = 2), Male (n = 2) Head: RIC(2) Body: gonadal sheath (non-neuronal) |
||||||
| SEROTONIN (5HT, 5-hydroxytryptamine) |
||||||
| Summary List of 5HT Neurons | ||||||
| DESCRIPTION | GENE NAME | DETECTION METHOD | LOCALIZATION | REFERENCES | ||
| 5HT | NA | HPLC + ED | Whole animal | Sanyal et al., 2004 | ||
| 5HT | NA | HPLC + ED | Whole animal | Wang et al., 2017 | ||
| 5HT | NA | Formaldehyde induced fluorescence (FIF)31 | NSM | Horvitz et al., 1982 | ||
| 5HT | NA | Glyoxylic acid induced fluorescence (GAIF)32 | NSM, ADF, AIM, RIH, HSN(h), VC4-5(h) | Duerr et al., 1999 | ||
| 5HT | NA | Antibody | NSM, ADF, AIM, RIH, HSN(h), VC4-5(h)33, CA1-4(m)34, CP1-6(m), RPAG(m)36, R1B(m), R3B(m), R9B(m) | Desai et al., 1988; Loer & Kenyon, 1993; Duerr et al., 1999; Jia & Emmons, 2006 | ||
| 5HT | NA | Antibody | ASG 23 | Pocock & Hobert, 2010 | ||
| 5HT | NA | Antibody | Extracellular 5HT-immunoreactivity observed following optogenetic stimulation of NSM, ADF [traditional criterion - stimulation-dependent release]. | Tatum et al., 2015 | ||
| 5HT | NA | Antibody | NSM, ADF, AIM4,35, RIH, URX(weak)35, I5(weak)35, CEM(m,weak)35, HSN(h), CP1-6(m), PGA(m)36, PVW(m, weak)37, R1B(m, weak), R3B(m), R9B(m) | Serrano-Saiz et al., 2017 | ||
Synthesis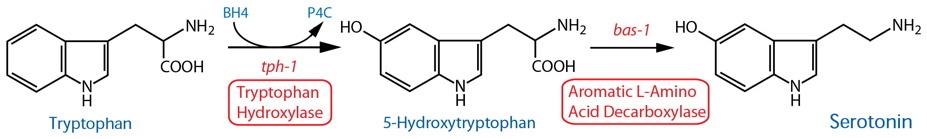 |
||||||
| Tryptophan Hydroxylase (TPH) | tph-1 |
Reporter transgenics | NSM, ADF, [AIM, RIH]39, HSN(h), CP1-6(m), R1B(m), R3B(m), R9B(m) | Sze et al., 2000 | ||
| TPH | tph-1 | Reporter transgenics | ASG38 | Pocock & Hobert, 2010 | ||
| TPH | tph-1 | Reporter transgenics (fosmid3) | NSM, ADF, HSN(h), CP1-6(m), R1B(m), R3B(m), R9B(m) | Serrano-Saiz et al., 2017 | ||
| TPH | tph-1 | Reporter transgenics | VC4-5(h)33 | Mondal et al., 2018 | ||
| Aromatic L-Amino Acid Decarboxylase (AADC)14 | bas-1 |
Reporter transgenics | NSM, ADF, [AIM, RIH]39, HSN(h), CP1-6(m), R1B(m), R3B(m), R9B(m); see also dopaminergic neurons |
Hare & Loer, 2004; Flames & Hobert, 2009 | ||
BH4 Cofactor Synthesis and Regeneration
|
||||||
| GTP |
cat-4 |
Reporter transgenics | NSM, ADF, HSN(h), CP1-6(m), R1B(m), R3B(m), R9B(m); see also Dopaminergic neurons | Sze et al., 2002 ; Flames & Hobert, 2009 | ||
| Pyruvoyl Tetrahydropterin Synthase (PTPS) | ptps-1 |
Reporter transgenics | NSM, ADF, HSN(h), VC4-5(h), other unidentified cells; see also Dopaminergic neurons | Zhang et al., 2014; Loer et al., 2015 | ||
| Pterin Carbinolamine Dehydratase (PCBD) | pcbd-116 |
Reporter transgenics | Various unidentified neurons and non-neuronal cells; see also Dopaminergic neurons | Zhang et al., 2014; Loer et al., 2015 | ||
| Quinoid Dihydropterin Reductase (QDPR) | qdpr-116 |
Reporter transgenics | NSM, ADF, other unidentified cells; see also Dopaminergic neurons | Zhang et al., 2014; Loer et al., 2015 | ||
| Transport | ||||||
| Vesicular Monoamine Transporter (VMAT) | cat-1 |
Antibody19 | NSM, ADF, AIM, male VNC & tail cells; see also Dopaminergic neurons | Duerr et al., 1999 | ||
| VMAT | cat-1 |
Reporter transgenics | NSM, ADF, AIM40, RIH, HSN(h), VC4-5(h), CP1-6(m), RPAG(m)36, R1B(m), R3B(m), R9B(m); see also DA, OA and TA neurons | Duerr et al., 1999; Nurrish et al., 1999; Duerr et al., 2001; Flames & Hobert, 2009 | ||
| VMAT | cat-1 |
Reporter transgenics | NSM, ADF, RIH, HSN(h), CP1-6(m), PGA(m, weak), PVW(male only, weak)37, R1B(m), R3B(m), R9B(m); see also DA, OA and TA neurons | Serrano-Saiz et al., 2017 | ||
| VMAT | cat-1 |
Uptake assay21 | NA - heterologous expression (CV-1 cells) | Duerr et al., 1999 | ||
| Serotonin Reuptake Transporter (SERT) | mod-5 |
Reporter transgenics | NSM, ADF, AIM, RIH, other neuronal and non-neuronal cells | Jafari et al., 2011; Barrière et al., 2014 | ||
| SERT | mod-5 |
Antibody | NSM, AIM | Jafari et al., 2011 | ||
| SERT | mod-5 |
Uptake assay41 | NA - heterologous expression (HEK293 cells) | Ranganathan et al., 2001 | ||
Catabolism |
||||||
| Serotonin N-Acetyltransferase (SNAT) | anat-1? | Enzymatic assay | Whole animal | Muimo & Isaacs, 1993 | ||
| Arylalkylamine N-Acetyltransferase (AA-NAT) |
anat-1?42 | Enzymatic assay | Whole animal | Migliori et al., 2012 | ||
| Monoamine Oxidase (MAO) | amx-125 | Reporter transgenics | ~30 head and tail neurons including ASJ, IL2, other amphid neurons; PHB, 3 other tail neurons | Filkin et al., 2007; Kaushal 2008 | ||
| MAO | amx-225 | Reporter transgenics | Intestine, neurons | Filkin et al., 2007 | ||
| MAO | amx-225 | Reporter transgenics | Pharynx, Intestine, vulval cells, rectal epithelial cells | Schmid et al., 2015 | ||
| MAO | amx-225 | Reporter transgenics | Pharyngeal muscle, intestine, NSM, other head neurons, vulval HSN(h), anus | Wang et al., 2017 | ||
Aldehyde Dehydrogenase (ALDH) |
alh-126 |
Reporter transgenics | Nervous system, including head neurons, neurons along body, PVT, intestine, head mesodermal cell, rectal gland cells, etc. | McKay et al., 200327 | ||
| Related Mutant Phenotypes / Other Supportive Evidence | ||||||
| TPH | tph-1 | Mutant lacks serotonin immunoreactivity (5HT-IR). | Sze et al., 2000 | |||
| GTPCH1 | cat-4 | Mutant lacks 5HT-IR (or is greatly reduced); see also Dopamine-related phenotypes. | Desai et al., 1988; Loer & Kenyon, 1993; Loer et al., 2015 | |||
| PTPS | ptps-1 | Mutant lacks 5HT-IR; see also Dopamine-related phenotypes. | Loer et al., 2015 | |||
| QDPR | qdpr-1 | Mutant has reduced 5HT-IR, especially combined with cat-4 reduction-of-function mutation; see also Dopamine-related phenotypes. | Loer et al., 2015 | |||
| PCBD | pcbd-1 | Mutant has reduced 5HT-IR, especially combined with cat-4 reduction-of-function mutation; see also Dopamine-related phenotypes. | Loer et al., 2015 | |||
| AADC | bas-1 | Mutant lacks 5HT-IR (or is greatly reduced); 5HT-IR rescued by exogenous 5HT but not 5HTP (5-hydroxytryptophan); see also Dopamine-related phenotypes. | Loer & Kenyon, 1993; Weinshenker et al., 1995; Sawin et al., 2000 | |||
| AADC | bas-1 | Age-related decline in bas-1 mRNA levels and reporter expression correlate with reduced NSM 5HT-IR. | Yin et al., 2014 | |||
| VMAT | cat-1 | Mutant lacks serotonin FIF in NSM processes, but shows increased FIF in somas; reduced 5HT-IR. | Horvitz et al., 1982; Loer & Kenyon, 1993 | |||
| VMAT | cat-1 | Expression of human VMAT1 or VMAT2 protein in mutant partially rescues behavioral phenotypes, and restores 5HT & dopamine-induced fluorescence (GAIF). | Duerr et al., 1999 | |||
| SERT | mod-5 | Mutant phenotype consistent with increased presynaptic serotonin, phenocopied by serotonin-specific reuptake inhibitors (SSRIs) such as fluoxetine, partially phenocopied by less-specific tricyclics such as imipramine; mutant is hypersensitive to exogenous serotonin. | Ranganathan et al., 2001 | |||
| SERT | mod-5 | Mutants lack 5HT-IR in AIM and RIH; fluoxetine or imipramine treatment reduces or eliminates 5HT-IR in AIM and RIH. In mutants, expression of mod-5 cDNA in AIM restores 5HT-IR; expression in other neurons causes ectopic 5HT-IR. | Kullyev et al., 2010; Jafari et al., 2011 | |||
| SERT | mod-5 | In tph-1; mod-5 double mutants, addition of 5HT fails to rescue loss of 5HT-IR. [Addition of 5HT in tph-1 single mutants rescues 5HT-IR in many neurons.] | Jafari et al., 2011 | |||
| MAO | amx-2 | Mutants have elevated 5HT levels; treatment with 5HIAA rescues altered vulval induction phenotype in mutant. | Schmid et al., 2015 | |||
| MAO | amx-2 | Mutants have elevated 5HT levels, and greatly reduced 5HIAA levels. Mutants in grk-2, which elevate AMX-2 levels, have greatly reduced 5HT levels, and greatly elevated 5HIAA levels. | Wang et al., 2017 | |||
| SUMMARY - Serotonergic Neurons (adult): Hermaphrodite (n = 11 or 13)38, Male (n = 20 or 22)38
By Class (with numbers and notes): By Body Region: Other possibly serotonergic neurons (not included in above totals): CA1-4(m)34, CEM(2, m)35, I535, PHB(2)43, URX35 |
||||||
| GAMMA-AMINOBUTYRIC ACID (GABA, gamma-aminobutyrate) |
||||||
| Summary List of GABA Neurons | ||||||
| DESCRIPTION | GENE NAME | DETECTION METHOD | LOCALIZATION | REFERENCES | ||
| GABA | NA | Antibody | AVL, DD1-6, DVB, RIS, RME, VD1-13 | McIntire et al., 1993b | ||
| GABA | NA | Antibody | ALA44, AVA45, AVB45, AVJ45, AVL, DD1-6, DVB, EF1-4(m), R2A(m), R6A(m), R9B(m), RIB, RIS, RME, SMDD/V45, VD1-13, GLR, hmc, muscle | Gendrel et al., 2016 | ||
| GABA | NA | Antibody | CP9(m), EF1-4(m), R2A(m, weak), R6A(m), R9B(m, weak) | Gendrel et al., 2016; Serrano-Saiz et al., 2017 | ||
|
||||||
| Glutamatic Acid Decarboxylase (GAD) | unc-25 |
Reporter transgenics | AVL, DD1-6, DVB, RIS, RME, VD1-13 | Jin et al., 1999 | ||
| GAD | unc-25 | Reporter transgenics (CRISPR-endogenous locus3) | AVL, DD1-6, DVB, EF1-4(m), RIB, RIS, RME, VD1-13 | Gendrel et al., 2016 | ||
| GAD | unc-25 | Reporter transgenics (CRISPR-endogenous locus3) | CP9(m) | Serrano-Saiz et al., 2017 | ||
| Transport | ||||||
| Vesicular GABA Transporter (VGAT) | unc-47 | Reporter transgenics | AVL, DD1-6, DVB, RIS, RME, VD1-13 | McIntire et al., 1997 | ||
| VGAT | unc-47 | Reporter transgenics | AVL, DD1-6, DVB , RIS, RME, SDQ(weak), SIAD, VD1-13 | Barrière & Ruvinsky, 2014 | ||
| VGAT | unc-47 | Reporter transgenics (fosmid3) | AVL, DD1-6, DVB , EF1-4(m), R6A(m), RIB, RIS, RME, SMDD/V, VD1-13 | Gendrel et al., 2016 | ||
| VGAT | unc-47 | Reporter transgenics (fosmid3) | CP9(m), EF1-4(m), R2A(m), R6A(m), R9B(m); Sex-shared cells with adult male only expression: ADF, AS10 or DA7, AS11, PDB, PHC, PVN; Loses expression in adult male: PQR; Cells that express unc-47 reporter but lack GABA-IR (possibly Glycinergic?): CA1-4(m)34, CEM(m), CP1-6(m), CP8(m), DVE(m), DVF(m), HOA(m), PDC(m), PGA(m), PVX(m), PVY(m), PVZ(m), R1A(m), R3A(m), R3B(m), R5A(m), R5B(m), R8A(m), R9B(m), PCB, PCC, SPC(m) | Serrano-Saiz et al., 2017 | ||
| 'VGAT Chaperone'46 | unc-46 |
Reporter transgenics | AVL, DD1-6, DVB , RIS, RME, VD1-13, plus a few unidentified neurons | Schuske et al., 2007 | ||
| 'VGAT Chaperone'46 | unc-46 | Reporter transgenics | AVL, DD1-6, DVB, RIS, RME, SIAD, VD1-13 | Barrière & Ruvinsky, 2014 | ||
| 'VGAT Chaperone'46 | unc-46 | Reporter transgenics (fosmid3) | AVL, DD1-6, DVB, EF1-4(m), R9B(m)47, RIB, RIS, RME,VD1-13 | Gendrel et al., 2016 | ||
| GABA Transporter (GAT) | snf-11 |
Reporter transgenics | AVL, [DD1-6,]48 DVB , [PVQ,]48 RIS, RME, [VD1-13]48 | Jiang et al., 2005 | ||
| GAT | snf-11 |
Reporter transgenics, antibody | AVL, DVB, RID, RIS, RME, 2 neurons in pharynx, 2 neurons in RVG, muscles (body wall, anal, uterine)48 | Mullen et al., 2006 | ||
| GAT | snf-11 |
Reporter transgenics (fosmid3) | ALA44, AVF, AVL, RIB, RME, VD12(m)49, GLR, hmc, muscle | Gendrel et al., 2016 | ||
| GAT | snf-11 |
Reporter transgenics (fosmid3) | CP9(m)49 | Serrano-Saiz et al., 2017 | ||
| GAT | snf-11 | Uptake assay50 | NA - heterologous expression (HRPE cells and Xenopus oocytes) | Jiang et al., 2005 | ||
| GAT | snf-11 | Uptake assay50 | NA - heterologous expression (Xenopus oocyte) | Mullen et al., 2006 | ||
Catabolism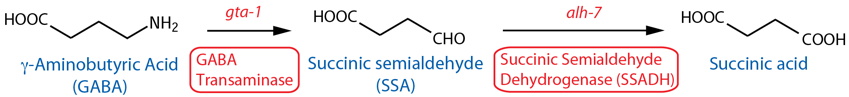
[Note: GABA catabolism has not been characterized in C. elegans. Although both GABA-T and SSADH (see below) are important in GABA catabolism in the mammalian brain (Tillakaratne et al., 1995), there is currently no evidence they serve the same functions in C. elegans (i.e., no revealing mutant phenotypes), but see below regarding recent gta-1 (GABA-T orthologous gene) expression data. Also, GABA-T transfers an amino group from GABA to α-Ketoglutarate to form Glutamate and SSA, so this reaction is also a potential source of glutamate. See also note26 regarding putative aldehyde dehydrogenases other than SSADH.] |
||||||
| GABA Transaminase (GABA-T) |
gta-1 |
Reporter transgenics | Body wall muscle, head neurons, unidentified cells | McKay et al., 200327; Meissner et al., 2011 | ||
| GABA Transaminase (GABA-T) |
gta-1 |
Reporter transgenics (fosmid3) | Ubiquitous51 | Gendrel et al., 2016 | ||
| Succinic Semialdehyde Dehydrogenase (SSADH) | alh-726 |
Reporter transgenics | Intestine, rectal gland cells, hypodermis, nervous system, head neurons | McKay et al., 200327 | ||
| Related Mutant Phenotypes / Other Supportive Evidence | ||||||
| GAD | unc-25 | Mutant lacks GABA immunoreactivity; mutant has 'shrinker' phenotype. | McIntire et al., 1993b | |||
| VGAT | unc-47 | Mutant has 'shrinker' phenotype characteristic of GABA loss of function, but elevated cellular GABA immunoreactivity. | McIntire et al., 1997 | |||
| VGAT Chaperone | unc-46 | Mutant has 'shrinker' phenotype. | McIntire et al., 1993b; Schuske et al., 2007 | |||
| GAT | snf-11 | Mutants show GABA-dependent aldicarb resistance. | Mullen et al., 2006 | |||
| GAT | snf-11 | GABA-dependent behaviors are not rescued by exogenous GABA in unc-25; snf-11 double mutants. | Mullen et al., 2006 | |||
| GAT | snf-11 | Mutants fail to accumulate GABA in cultured embryonic cells. | Mullen et al., 2006 | |||
| SUMMARY - GABAergic Neurons (adult): Hermaphrodite (n = 32), Male (n = 43)
By Class (with numbers and notes): By Body Region: Other possibly GABAergic neurons (not included in above totals): AVA, AVB, AVJ; possible 'GABA clearance' neurons: ALA, AVF |
||||||
| GLUTAMATE (Glu, Glutamic Acid) |
||||||
| Summary List of Glu Neurons | ||||||
| ||||||
| DESCRIPTION | GENE NAME | DETECTION METHOD | LOCALIZATION | REFERENCES | ||
| Transport | ||||||
| Vesicular Glutamate Transporter (VGluT) | eat-452 |
Reporter transgenics | M3, I5, ADA, ALM, ASH, ASK, AUA, AVM, FLP, IL1, LUA, OLL, OLQ, PLM, PVD, PVR | Lee et al., 1999 ; Mano et al., 2007 | ||
| VGluT | eat-452 |
Reporter transgenics | Many head neurons, including ADL, AFD, AIB, AIM, AIZ, ASH, ASE, ASG, ASK, AUA, AVA, AVE, AWB, AWC, RIA, etc. | Ohnishi et al., 2011 | ||
| VGluT | eat-452 |
Reporter transgenics (fosmid3) | M3, MI, I2, I5, ADA, ADL, AFD, AIB, AIM, AIZ, ALM, AQR, ASE, ASG, ASH, ASK, AUA, AVM, AWC, BAG, DVC, FLP, IL1, LUA, OLL, OLQ, PHA, PHB, PHC, PLM, PQR, PVD, PVQ, PVR, RIA, RIG, RIM, URY, 20 more male-specific cells (see below) | Serrano-Saiz et al., 2013 | ||
| VGluT | eat-452 |
Reporter transgenics (fosmid3) | Male-specific cells only: CP0(m, weak), CP5(m, weak), CP6(m, weak), CP7(m), HOA(m), PCA(m), PVV(m), R2B(m, weak), R5A(m), R6B(m, weak), R9A(m, weak) | Serrano-Saiz et al., 2017 | ||
| VGluT | vglu-253 | Reporter transgenics (fosmid3) | AIA, skin cells | Serrano-Saiz et al., 2020 | ||
| Plasma Membrane Glutamate Transporter (PmGluT) | glt-154 |
Reporter transgenics | Muscle, hypodermis | Mano et al., 2007 | ||
| PmGluT | glt-3 |
Reporter transgenics | Excretory canal cell, pharynx | Mano et al., 2007 | ||
| PmGluT | glt-455 |
Reporter transgenics | AUA, RIA, IL2 | Mano et al., 2007 | ||
| PmGluT | glt-5 |
Reporter transgenics | Pharynx | Mano et al., 2007 | ||
| PmGluT | glt-6 |
Reporter transgenics | Excretory canal cell, pharynx marginal cells | Reported27 in Mano et al., 2007 | ||
| PmGluT | glt-7 |
Reporter transgenics | Excretory canal cell | Mano et al., 2007 | ||
| Related Mutant Phenotypes/ Other Supportive Evidence | ||||||
| VGluT | eat-4 | Mutant has pharyngeal phenotypes similar to glutamate receptor avr-15 mutant, but pharynx responds normally to exogenous Glu, indicating presynaptic function. | Dent et al., 1997 | |||
| VGluT | eat-4 | Mutant phenotypes (hyperactive foraging, reduced pharyngeal pumping rate) are rescued by expression of human VGluT via eat-4 promoter. | Lee et al., 2008 | |||
| PmGluT | glt-1 - glt-7 | Individual glt mutants have increased Glu-dependent behaviors. | Mano et al., 2007 | |||
| PmGluT | glt-3, glt-4, glt-6 glt-3, glt-4, glt-7 |
Triple glt mutants have strongly increased Glu-dependent behaviors. | Mano et al., 2007 | |||
| SUMMARY - Glutamatergic Neurons (adult): Hermaphrodite (n = 79), Male (n = ~98)
By Class (with numbers and notes): By Body Region: |
||||||
Neurons currently lacking classical neurotransmitter assignments (list taken from Serrano-Saiz et al., 2017) Sex-shared neurons: ASI, AVF, AVH, AVJ, AVK, AWA, BDU, CAN20, PVM, PVT, PVW [only in hermaphrodites], RID, RIP, RMG, I4, and I6 |
||||||
Notes
2 Because of the genomic structure of the unc-17-cha-1 locus, it is likely that the genes are co-expressed in the same neurons. However, expression of cha-1 was confirmed only for the unc-17 -expressing neurons identified in the Duerr et al., 2008 study. 3 Most reporter transgenics to date have been made with a few kbp of upstream genomic sequence fused to the reporter (transcriptional, or translational near the N-terminus), and therefore may lack important regulatory elements (distant upstream, within coding, and downstream). Larger fosmid-based reporters typically have 35-40 kbp of genomic sequence, both upstream and downstream, with the reporter fused to the full length coding sequence at the C-terminus, or separated from the gene of interest by an SL2-splice site creating an artificial operon (e.g., see Dolphin & Hope, 2006; Sarov et al., 2006; Tursun et al., 2009). Therefore, transgenics with these fosmid (or BAC) constructs may be more likely to reproduce the expression pattern of the endogenous locus. Issues with both small fusion reporter constructs and fosmid reporters that are extrachromosomal or inserted into ectopic genomic locations may also progressively be resolved with CRISPR-generated knock-in reporters of various types inserted as single copies into the endogenous locus. 4 AIM neurons are dual-transmitter glutamatergic/serotonergic in hermaphrodites (Serrano-Saiz et al., 2013); in males, AIM neurons are glutamatergic/serotonergic until L3, then switch to a cholinergic/serotonergic dual-transmitter phenotype (Pereira et al., 2015). 5 These cells express unc-17 but not cho-1 (Pereira et al., 2015). 6 DVA was originally (mis)identified as DVC by Duerr et al., 2008. 7 Pereira et al., 2015 originally identified 2 male-specific tail neurons as DVE & DVF; Serrano-Saiz et al., 2017 revised this identification to be DX1/2. 8 Expression of cho-1 is strong in RIB, but unc-17 expression is very weak (Pereira et al., 2015). 9 VC4-5 express unc-17 but not cho-1 (Pereira et al., 2015); VC4-5 also release serotonin (Duerr et al., 1999). 10 Uptake was Na+-dependent, and blocked by 1 μm HC3 (criterion for high-affinity uptake) (Okuda et al., 2000). 11 Staining with acetylthiocholine (Method: Karnovsky & Roots, 1964). 12 Although the genes ace-4 and ace-3 are coexpressed in a bi-cistronic operon (controlled by a single promoter upstream of ace-4), the level of ace-4 mRNA is exceedingly low, and there is no detectable ACE-4 protein or enzymatic activity present in worms (Combes et al., 2000). Also, there is no known ace-4-orthologous gene in nematodes outside of genus Caenorhabditis; only an ace-3 gene is present. . 13 Spicule socket cells SPSo ('non-neuronal' support cells) comprise 2 syncitial cells with 4 nuclei each that likely release dopamime to promote sperm release, acting much like neurons (LeBouef et al., 2014). Interestingly, SPSo cells do not appear to express DAT-1, and SPSo-mediated behavior is not rescued by exogenous dopamine (LeBouef et al., 2014). 14 AADC is also known as 5HTP Decarboxylase or Dopa Decarboxylase (DDC); in animals, the enzyme generally has broad substrate specificity, catalyzing both serotonin and dopamine synthesis (reviewed by Zhu & Jorio, 1995). 15 BH4 is also required for the function of other aromatic amino acid hydroxylases such as phenylalanine hydroxylase (pah-1), and the lipid metabolic enzyme alkylglycerol monooxygenase (agmo-1); therefore, expression of BH4 synthesis and regeneration genes is not neuron-specific. They are highly expressed in the hypodermis, and also in the intestine (Loer et al., 2015). BH4 is also required for nitric oxide synthase (NOS) function; however, NOS has not been identified in C. elegans (Gusarov et al. 2013). 16 Reporters for these BH4 synthesis and regeneration genes to date have shown limited expression in identified dopaminergic and serotonergic neurons (Zhang et al., 2014; Loer et al., 2015), despite mutant phenotypes indicating function in those cells promoting serotonin and dopamine synthesis (Loer et al., 2015). 17 QDPR is also known as Dihydropteridine reductase (DHPR). 18 Vesicular monoamine transporter (VMAT) is used in common by all monoaminergic neurons to transport the neurotransmitter into synaptic vesicles for release (Eiden & Weihe, 2011). In C. elegans, this includes neurons using dopamine, tyramine, octopamine, serotonin and perhaps one or more as yet unidentified monoamines20. 19 VMAT colocalizes with synaptic vesicles (Duerr et al., 1999). 20 Expression of a cat-1/VMAT fosmid (but no other monoaminergic markers) may indicate CAN uses an unidentified monoamine (Serrano-Saiz et al., 2017). 21 VMAT's relative affinity for monoamine substrates: dopamine ~ tyramine > serotonin > norepinephrine ~ octopamine > histamine (Duerr et al., 1999). 22 DAT expressed in human cells mediates Na+ and Cl--dependent uptake of dopamine better than norepinephrine or other transmitters. Transport by DAT was blocked by tricyclics (especially imipramine) and other monoamine transport inhibitors (Jayanthi et al., 1998). 23 DAT mediates Na+ and Cl--dependent uptake of dopamine in both heterologous (tsA-201) and cultured native C. elegans cells, and shows electrogenic activity (Carvelli et al., 2004). 24 Among the named C. elegans comt genes, four encode proteins orthologous to mammalian COMT-domain containing protein 1 (COMT-D1), and are most similar to bacterial and plant known and putative O-methyltransferases, including Caffeic Acid O-Methyltransferase (also abbreviated COMT) and Caffeoyl CoA O-Methyltransferase (CCoAOMT, Ferrer et al., 2005). It is plausible that one or more of the worm COMT proteins could act on a catechol-containing substrate based on similarity to CCoAMTs - i.e., the substrate caffeoyl CoA has a catechol structure that is methylated on the 3-hydroxyl by CCoAMT. The worm COMT proteins, however, have very limited or no significant sequence homology to mammalian COMTs except among S-adenosylmethionine binding residues (SAM aka AdoMet, the methyl donor) found in all AdoMet-dependent methyltransferases (e.g., see Martin & McMillan, 2002). Currently, there is no evidence suggesting comt gene function in neurotransmitter metabolism; no informative mutant phenotypes (by RNAi) and no expression patterns have been reported. 25 Among the amx gene encoded proteins, the AMX-2 predicted protein is most similar to a mammalian monoamine oxidase (MAO-A), and has been shown to metabolize serotonin (Schmid et al., 2015). AMX-1 may be a histone demethylase (orthologous to lysine-specific histone demethylases). The protein encoded by amx-3 (not listed) is most similar to polyamine and spermine oxidases; no expression pattern has been reported to date. 26 There are 13 identified alh genes that encode proteins orthologous or highly similar to mammalian aldehyde dehydrogenases (ALDHs). Those shown here have relevant reporter expression patterns. To date, there are no expression patterns reported for alh-2, alh-3, alh-4, alh-5, alh-11, and alh-12; alh-8 is expressed in muscle mitochondria (Meissner et al., 2011); alh-9 is expressed only in hypodermis, during embryogenesis (Mounsey et al., 2002); alh-13 is expressed in the adult intestine (McKay et al., 2003). See also GABA catabolism regarding alh-7 / SSADH. Aldehyde dehydrogenases convert a wide array of both endogenous and exogenous aldehydes to carboxylic acids in amino acids, biogenic amines, carbohydrates, lipids, etc. In humans, the same ALDH (ALDH1A1) that converts DOPAL to DOPAC in dopminergic neurons elsewhere converts retinal to retinoic acid, and oxidizes the ethanol metabolite acetaldehyde (Marchitti et al., 2008). ALDH1A1 is also capable of mediating GAD-independent GABA synthesis in mammalian brain via an alternative pathway from putrescine (Seiler & Al-Therib, 1974; Kim et al., 2015). 27 Found in Hope Expression Pattern DB (Reference: Hope et al., 1996). 28 Method of P. D. Evans, 1978. 29 The uterine uv1 cells appear to be neuroendocrine. They express neurosecretory proteins and neuropeptides, contain neuroscretory vesicles, and likely release tyramine to inhibit egg laying (Alkema et al., 2005 and references therein). 30 Neither R8A or R8B express a cat-1/VMAT fosmid, making it unclear whether they actually use tyramine as a neurotransmitter (Serrano-Saiz et al., 2017). 31 FIF for serotonin in C. elegans is weak and extremely labile (Horvitz et al., 1982), which likely explains why it has been seen only in the NSM neurons. 32 GAIF method of de la Torre, 1980; comparable to FIF, but apparently more robust in C. elegans. 33 5HT-IR in VC4-5 can be weak and unreliable (Duerr et al., 1999), and/or varies with antiserum and staining protocol. Although at least one smaller tph-1 reporter construct does show expression, Serrano-Saiz et al., 2017 did not observe 5HT-IR or cat-1/VMAT fosmid reporter expression in VC4-5. 34 5HT-IR in CA neurons (CA1-4) is rare (Loer & Kenyon, 1993), but is not reported by others (e.g., Serrano-Saiz et al., 2017). CA neurons also do not express other serotonergic marker genes. Strong 5HT-IR in likely CA homologs is observed in other nematodes, including other Caenorhabditis species (Loer & Rivard, 2007). 35 Serrano-Saiz et al., 2017 suggest that AIM, CEM, I5 and URX are '5HT clearance' neurons - they saw no expression of tph-1 or cat-1/VMAT in the cells. Unlike the other cells, which are weakly 5HT immunoreactive, AIM stains strongly and reliably with serotonin antibodies. 36 The male-specific, unpaired 'right preanal ganglion' (RPAG) neuron was first identified as either PDC or PGA (Loer & Kenyon, 1993); it is now known to be PGA (Serrano-Saiz et al., 2017). 37 5HT-IR and cat-1/VMAT fosmid expression in PVW is sexually dimorphic, found only in the adult male (Serrano-Saiz et al., 2017). 38 ASG is glutamatergic, but 5HT-IR and tph-1 reporter expression appear after exposure to hypoxic conditions (Pocock & Hobert, 2010). 39 Expression of tph-1 and bas-1 in AIMs and RIH is rare with most reporters; these cells may be '5HT-absorbing' via MOD-5/SERT (and serotonin-releasing) rather than 5HT-synthesizing neurons (Kullyev et al., 2010; Jafari et al., 2011). No expression was observed in the RIH and AIM (and VC4-5) neurons with tph-1 fosmid reporter (Serrano-Saiz et al., 2017). 40 Reported CAT-1/VMAT antibody staining in AIM (Duerr et al., 1999) may actually be in the tyraminergic RIM, based on a cat-1 fosmid reporter that otherwise fully replicates previous antibody staining (Serrano-Saiz et al., 2017). 41 MOD-5 expressed in mammalian cells mediates Na+-dependent uptake of 5HT but not dopamine, histamine, norepinephrine, GABA, Glu, or Gly. Uptake is blocked by SSRIs, tricyclics, and non-specific monoamine transport inhibitors (Ranganathan et al., 2001). 42 AA-NAT activity may be catabolic for serotonin and/or synthetic for melatonin. Although there are no clear orthologs of hydroxyindole-o-methyltransferase (HIOMT) in C. elegans, synthesis of melatonin, and a reporter expression pattern for a very weak homolog (Y74C9A.3/homt-1) has been reported (Tanaka et al., 2007; Migliori et al., 2012). 43 I5 and PHB are glutamatergic (Serrano-Saiz et al., 2013), and may also be serotonergic (Sawin et al., 2000). 44 Cells / tissues likely mediate GABA clearance and/or recycling; they do not express unc-47 (VGAT) (Gendrel et al., 2016). 45 Anti-GABA staining in AVA, AVB and AVJ is weak; the cells express no other GABA-related marker genes (Gendrel et al., 2016). 46 UNC-46 is a BAD-LAMP-related protein required for trafficking UNC-47/VGAT to synaptic vesicles at synapses, specifically in GABAergic neurons in worms. Rescuing UNC-46-GFP fusion proteins co-localize with synaptic varicosities (Schuske et al., 2007). 47 Expression is weak and inconsistent. 48 There are discrepancies between the snf-11 expression patterns reported by Jiang et al. (2005) and those reported by Mullen et al. (2006); the Jiang et al observations are likely due to an error in promoter construction (see Mullen et al., 2006). 49 Male-specific expression of snf-11 in VD12 reported in Gendrel et al., 2016 is instead expression in the male-specific CP9 (Serrano-Saiz et al., 2017). 50 SNF-11 expressed in Xenopus oocytes (Jiang et al. 2005; Mullen et al., 2006) or in mammalian HRPE cells (Jiang et al. 2005) mediates Na+- and Cl--dependent high affinity uptake of GABA; uptake is blocked by GAT inhibitors nipecotic acid and SKF89976A (Mullen et al., 2006). 51 Ubiquitous expression of gta-1 (single GABA-T orthologous gene) is consistent with a role beyond GABA metabolism (Gendrel et al., 2016). 52 Lee et al., 1999 reported eat-4 expression in NSM, AVJ, and IL2, and Ohnishi et al., 2011 reported eat-4 expression in the AVA, AVE, SIB, RMD and ASJ neurons; however, subsequent analysis with the same transgenic reporter constructs did not replicate those cell IDs (Serrano-Saiz et al., 2013). 53 Reporter expression localizes to vesicular structures in the soma, indicating VGLU-2 may not be involved in synaptic vesicle function; however, vglu-2 mutants have AIN-associated olfactory behavior defects indicating function. In C. elegans alone (not other sequenced Caenorhabditis species), there is a degenerate duplicate gene, vglu-3, for which reporters show no expression (Serrano-Saiz et al., 2019). 54 There is no glt-2 (the sequence originally named glt-2 was found to be a splice variant of glt-1). Although all glt genes are listed here, it seems unlikely that all are involved in neuronal glutamate use. 55 glt-4 may be a primarily presynaptic neuronal GluT. |
||||||
Abbreviations
|
||||||
Acknowledgements and General References
|
||||||
|
||||||
|
|